>Corresponding Author : Jawhar Gharbi1
>Article Type : Review Article
>Volume : 1 | Issue : 1
>Received Date : 2 November, 2021
>Accepted Date : 16 November, 2021
>Published Date : 20 November, 2021
>DOI : https://doi.org/10.54289/JVVD2100103
>Citation : Hamrita B, Almalki MA, Hafi F, M’hadheb MB, Gharbi J (2021) Emerging Human Coronaviruses: Molecular Biology and Vaccine Challenges J Virol Viral Dis 1(1). doi https://doi.org/10.54289/JVVD2100103
>Copyright : © 2021 Hamrita B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Review Article | Open Access | Full Text
1Virology & Antiviral Strategies Research Unit UR17ES30 (ViroBiotech), Higher Institute of Biotechnology, University of Monastir, BP 74, Tahar HADDED Street, Monastir 5000, Tunisia
2Department of Biological Sciences, College of Science, King Faisal University, P.O. Box 400, Al-Ahsa 31982, Saudi Arabia
3Department of Anesthesiology and Surgical Intensive Care Unit, FattoumaBourguiba University Hospital, BP 15, Avicenne street, Monastir 5000, Tunisia
*Corresponding author: Jawhar Gharbi, Department of Biological Sciences, College of Science, King Faisal University, P.O. Box 400, Al-Ahsa 31982, Saudi Arabia
Abstract
COVID-19 is a new public health crisis caused by the novel respiratory pathogen SARS-CoV-2. It is one of the most significant pandemic events in recent history. The SARS-CoV-2 Beta corona virus was transmitted to humans in the end of 2019 by unknown intermediary host from bats in Wuhan, Hubei province (China). It marked the third major coronavirus source of disaster in the 21stcentury.The three last severe respiratory tract infections caused by the SARS-CoV-1, MERS-CoV and SARS-CoV-2 caused high human mortality. Viral genomic sequencing and investigations and the development of advanced vaccine strategies are expected to give us more information on these emerging pathogens and controlling them in the future. The aim of this review is to summarize updated information regarding these emerging human coronaviruses to understand their molecular and structural biology, transmissions and potential vaccine approaches actually developed against the SARS-CoV-2.
Keywords: Coronaviruses, SARS-CoV-1, MERS-CoV, SARS-CoV-2, COVID-19, Vaccines.
Abbreviations: COVID-19: Coronavirus Disease 2019; HCoVs: Human Coronaviruses; SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; SARS-CoV-1: Severe Acute Respiratory Syndrome Coronavirus 1;MERS-CoV: Middle East Respiratory Syndrome Coronavirus; S: Spike protein; RBD: Receptor-Binding Domain; RBM: Receptor-Binding Motif; ACE2: Angiotensin-Converting Enzyme 2;WHO: World Health Organization; ICTV: International Committee on Taxonomy of Viruses; ORFs: Open Reading Frames; NSP: Nonstructural Protein; UTR: Untranslated Region; CTL: Cytotoxic T Lymphocytes; APC: Antigen Presenting Cells; CEPI: Coalition for Epidemic Preparedness Innovations.
1. Introduction:
Coronaviruses (CoVs) belong to the viral order “Nidovirales” and has four genera (alpha, beta, gamma, and delta coronaviruses) [1]. It can provoke many diseases in several species of animals and humans. Animal species that are infected with CoVs include horses, camels, dogs, cats, birds, bats, rabbits and various of other wild animals. The CoVs infecting humans (HCoVs) belong to alpha and beta coronaviruses [1]. Actually, there are seven HCoVs in the world: the alpha coronaviruses infecting humans are HCoV-229E and HCoVNL63 and the beta coronaviruses infecting humans are HCoV-HKU1, HCoV-OC43, MERS-CoV,
SARS-CoV-1, and actually SARS-CoV-2 [2]. Among these seven strains, three strains proved to be highly pathogenic (SARS-CoV-1, MERS-CoV and SARS-CoV-2) which caused epidemic and pandemic of severe CoVs diseases [2]. HCoVs infections are widely associated with severe respiratory diseases, headache, diarrhea, cough, high fever, high inflammation, fatigue and hemoptysis [2,3]. Understanding the origin of HCoVs and their symptoms will be helpful for disease evolution, development of new target drugs and prevention of further epidemics. Before the first outbreak of SARS-CoV-1, a limited number of CoVs were known to be circulating in humans, causing illnesses such as the common cold and infrequently children affected and deaths. The world experienced of CoVs infections that threaten global pandemic in 2002-2003 by SARS-CoV-1and in 2012 by MERS-CoV. In both cases, the causative agents SARS-CoV-1 and MERS-CoV, respectively were newly identified in the genus Beta corona virus with zoonotic origin [1].
At the end of 2019, an outbreak of another HCoVs that causes respiratory-related illness was reported in Wuhan, Hubei province in China. The disease is named by the WHO “The Corona Virus Disease-2019 COVID-19” [2,4]. WHO has declared this disease a global health emergency at the end of January 2020.TheInternational Committee on Taxonomy of Viruses (ICTV) named the novel coronavirus causing the disease SARS-CoV-2. Subsequently, the experts of the ICTV termed it the SARS-CoV-2 as it is very similar to the one that caused the SARS precedent outbreak (SARS-CoV-1) [2]. The virus was identified, and its genome is fully sequenced in 7th January, 2020.The SARS-CoV-2 until now is less pathogenic as compared to SARS-CoV-1 and MERS-CoV. Since its first reported case in late 2019, the infection has spread to other regions in China and to more than 266 countries. The transmission and the mortality highly emerged in some countries as USA, Brazil, Russia, Spain, Italy and UK. It has been proven that SARS-CoV-2 is transmitted by inhalation or contact with infected droplets and the incubation period is up to 2 or 3 weeks but in most cases is significantly shorter [5]. According to the WHO, viral diseases continue to emerge and represent a serious issue to public health. The symptoms are usually fever, cough, sore throat, breathlessness, fatigue, and malaise. As there is no actually defined therapy available, we review in this paper the molecular and structural biology of these viruses and we analyses the development of new vaccine strategies as preventive approaches against the virus and the COVID-19 disease.
2. Emerging human coronavirus infections
2.1. SARS-CoV-1
The story of the CoVs infection began in November 2002 [6]. The first known case of SARS-CoV-1 emerged in Southern China and spread around the globe in the end of 2002. The reservoir of SARS-CoV-1 is unknown. The bats and subsequent spread to Himalayan palm civets are hypothesized [6]. From, February 2003, more than 300 cases had been reported and the majority of which were in medical staff [7]. Individuals who were infected with SARS-CoV-1 and subsequently travelled and emerged the outbreak to other countries. In March 2003, the WHO established a network of laboratories to determine the causative agent of the SARS infection [7]. In April 2003, a remarkable global effort led to the identification of SARS coronavirus. In July 2003 this virus infecting at least of 8,096 reported cases, with 10 % of case fatality rate (774 cases) in 27 countries by the first source of disaster in the 21st century SARS-CoV-1outbreak [7].
2.2. MERS-CoV
Almost a decade later, exactly in June 2012, the second source of disaster in the 21st century is displayed when a man in Saudi Arabia died of acute pneumonia and renal failure [8]. The etiology of this pathology is attributed to a virus belonging to the CoVs family. A novel bat corona virus was isolated and emerged in the human population, named Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [8,9]. Like SARS-CoV-1, MERS-CoV emerged as a result of zoonotic introduction to the human population, and it has demonstrated that dromedary camels as the most probable reservoir for MERS-CoV [9]. In contrast, to the SARS-CoV-1epidemic, which was contained within one year, MERS-CoV still continues circulating after its first identification and cause outbreaks [10]. In fact, infections have already been reported in more than 27 countries across the Middle East, Europe, North Africa and Asia [10]. In April 2012, a cluster of cases of severe respiratory disease had occurred in Jordan hospital and was retrospectively diagnosed as MERS-CoV, and a cluster of three cases of MERS-CoV in the UK was identified in September 2012 [10,11]. MERS-CoV continued to emerge to countries outside of the gulf territory as a result of travel of infected persons often. In May 2015, a single person returning from the Middle East started a nosocomial outbreak of MERS-CoV in South Korea that involved other patients. Novel cases have since been detected in Tunisia, Germany, France, Italy, the USA and UK [12-15]. Finally, in March 2017, 1905, laboratory-confirmed cases of MERS-CoV in 27 countries have been reported to the WHO including 677 deaths.
2.3. SARS-CoV-2
Since December 2019, a novel CoV named SARS-CoV-2emerged and displayed the third disaster in the 21st century [2]. In Chinese Wuhan, capital city of Hubei province, many cases started presenting to local hospitals with severe pneumonia [2]. The surveillance system was activated and on December 31, 2019, China notified the outbreak to the WHO. On 7th January, the virus was identified as a novel bat CoVs and presented a high resemblance with the SARS-CoV-1. SARS-CoV-2 likely originated in bats but might have been amplified in an intermediate host [16]. The first death case was reported in Wuhan on 11th January 2020 [17]. From January 23rd, Wuhan population was placed under lock-down with restrictions of entry and exit from the region. Despite all that, virus outbreak was emerged to other places of Hubei province [18]. SARS-CoV-2 virus was identified within 28 days of their spread in China. This is fast emergence compared to the time taken to identify SARS-CoV-1 reported in Chinese Foshan, Guangdong Province. The number of cases increasing exponentially, some of which did not have exposure to the live animal, suggestive that human-to-human transmission was occurring. Study conducted in Hong Kong University has provided the first concrete evidence for human-to-human transmission of SARS-CoV-2. RNA tests can confirm the diagnosis of COVID-19 cases with real-time RT-PCR, and it is considered an effective method for confirming the diagnosis in clinical cases of COVID-19 [18]. All SARS-CoV-2 cases in countries outside China were reported in those with no history of travel to China confirming that local human-to-human transmission was occurring in these countries. The most potential risk for the spread of COVID-19 worldwide is related to travel that is causing the regional and global spread of the disease [2]. In addition, airports in many countries put in screening mechanisms to detect symptomatic people returning from China and placed them in isolation and testing them for SARS-CoV-2. Soon, it was apparent that the infection could be transmitted from asymptomatic people. Therefore, countries who evacuated their citizens from Chinese Wuhan placed all people in strict confinement of 14 days [5].
COVID-19 has already evolved into a pandemic spreading rapidly worldwide and cases continued to increase exponentially [2]. Data provided in the end of October 2021, a total number of more than 250 million reported cases worldwide of COVID-19, including more than 4.8 million deaths. The majority of cases have been reported from Wuhan to USA, Brazil, Russia, France, Spain, United Kingdom, Italy, and India. World numbers are possibly an underestimate of the infected and dead due to limitations of surveillance and testing. The most up-to-date source for the epidemiology of this emerging pandemic can be found daily at the following sources: The WHO Novel Coronavirus (COVID-19) Situation Board and The Johns Hopkins Center for Systems Science and Engineering site for Coronavirus Global Cases COVID-19, which uses openly public sources to track the spread of the pandemic.
3. Emerging human coronaviruses: Structure and transmission
The three last emerged CoVs present the largest genome among RNA viruses [10]. CoVs consisting of a single-stranded positive-sense RNA molecule capped, and poly-adenylated RNA genomes (~30 kb in length) which encodes for both structural and non-structural proteins [19]. CoVs are belongs to Coronaviridae family of the Ortho coronavirinae sub family which can cause illness in birds, mammals and humans [9]. The Structural Proteins (sp) known of CoVs are (S) Spike protein, Membrane (M) protein, Envelop (E) protein, and Nucleocapsid (N) protein [1] (Figure 1). The genome is similar to cellular mRNAs, as it contains 5′-capped and 3′ polyadenylated ends. The RNA of CoVs has seven genes that are conserved in the order: ORF1a, ORF1ab, S, ORF3, E, M, N in 5' to 3' direction [20]. The 5′-terminal constituting the 2/3part of the RNA genome is covered by two overlapping open reading frames (ORFs): ORF1a/ab, which produces the 2 viral replicase proteins that are polyproteins (PP1a and PP1ab) [21]. Sixteen mature Nonstructural Proteins (nsp) derive from further processing of these two PPs [22-23] (Figure 2). These nsp take part in different viral functions including the formation of the replicase transcriptase complex (RTC). Most of them being involved in viral genome replication and sub genomic mRNA synthesis [22]. All sp and nsp may serve as drug design targets.
Total nsp have various roles like suppression of gene expression by nsp1/2, nsp3 participate in the multidomain complex formation, nsp5 is the main protease [24], nsp4/6 which are transmembrane (TM) proteins [25], nsp7 and nsp8 may bind and regulate the core enzyme of the RTC [26] and nsp9 as RNA-binding protein for participate in viral spreading [27]. The nsp10 is implicate in the activation of the replicative enzyme and the nsp12 present RNA-dependent RNA polymerase activity [28]. A helicase activity of nsp13 and nsp14 insured exoribonuclease activity [25]. Finally, nsp 15 and nsp 16 present endoribonuclease and methyltransferase activities [25]. The 16nsp have an important role in the CoVs emergence by their implications roles in replication and transcription [25]. Another important envelop-associated protein which is expressed by only some strains of CoV is the HE protein [29]. The RNA genome of CoV is packed in the nucleocapsid protein (N) and further covered with envelope (E) [22].
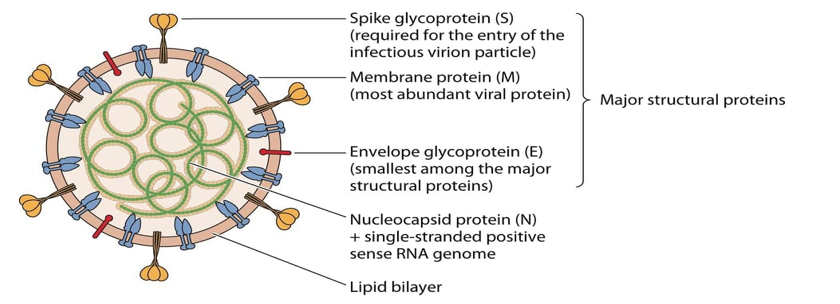
Figure 1: EmergingHumanCoVs viral particle structure with the major Structural Proteins (sp) [1].
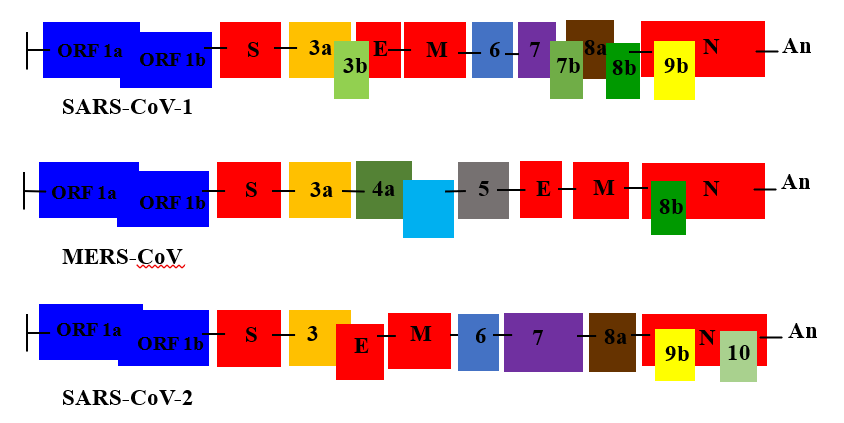
Figure 2: Emerging Human CoVs Genome structures. ORF 1a and ORF 1ab represent the two thirds of the genome, they are encoding components of the Replication/Transcription Complex RTC. The remaining one third of the genome encoding the structural and accessory proteins. Genes encoding major structural proteins sp: Spike (S), Membrane (M), Envelope (E) and Nucleocapsid (N) are indicated [30].
3.1. SARS-CoV-1
The SARS-CoV-1 genome has 29727 nucleotides consists of 11 putative open reading frames (ORFs) in 9 mRNA transcripts [31]. ORF1a and ORF1ab containing about two-thirds of the genome, both encode large polyproteins [32]. The genome encodes four major structural proteins (spike (S), envelope (E), membrane (M), and nucleocapsid (N) that play a major role in virus entry and replication in the host cell.
Additionally, SARS-CoV-1also has several small non-structural ORFs that are found between the S and E genes and between the M and N genes [33] (Figure 2 and Table 1). This virus is different from other human CoVs for many reasons, such as the short anchor of the S protein, the number and location of ORFs, and the presence of only one copy of PLpro (papain-like protease). The SARS-CoV-1 seems remarkably invariant comparing to the SARS-CoV-2 because of the proof-reading capacity of nsp14.The genome sequences of various SARS-CoV-1 patients have not revealed any changes [34]. SARS-CoV-1brought about strong immunological responses contributed to viral protection and pathogenesis. Transmission is through droplet infection and close person-to-person contact [2,22]. It can also spread through sweat, stool, urine, and respiratory secretions. When virus enters into the body, it binds to the primary target cells such as pneumocytes, and thereby establishing a cycle of infection and replication [2,22]. Other target cells of SARS-CoV-1 are epithelial renal tubules, tubular epithelial cells of kidney, enterocytes, immune cells, and cerebral neuronal cells. SARS-CoV-1 attaches to the target cells with the help of spike protein-host cell protein interaction (Angiotensin Converting Enzyme-2: ACE-2) [7]. Protein spike glycoprotein initiate SARS-CoV-1entry into cells by binding to ACE2 receptors (318-510 amino acids), followed by conformational changes resulting in membrane fusion. After the receptor recognition, the virus genome with its nucleocapsid enter into the cytoplasm of the target cells [22]. The primary function of a nucleocapsid protein is to form helical ribonucleoprotein complexes with viral RNA [22]. In addition to comprising the core structure of the SARS-CoV-1 virion, such complexes are involved in multiple functions such as transcription, replication, and virus packaging [35]. Minor alterations in the S protein can affect tissue and coronavirus virulence [22].
The S spike protein of SARS-CoV-1 containing (1255-amino-acids-length) 4 parts: a signal peptide located at the N terminus (1 to 12 amino acids), an extracellular domain (13 to 1195 amino acids), a transmembrane domain (1196 to 1215) amino acids, and an intracellular domain from amino acids 1216 to 1255 [36]. Proteases such as trypsin and cathepsin L cleave the SARS-CoV-1S protein into two subunits, the S1 and S2 subunits [31,36]. A minimal receptor-binding domain (RBD) located in the S1 subunit (318-510 amino acids) can combine with ACE2, the host cell receptor. The RBD displays a concave surface during interaction with the receptor [29]. The entire receptor-binding loop, known as the receptor-binding motif (RBM) (424-494 amino acids) is located on the RBD and is responsible for complete contact with ACE2 [29]. Importantly, two residues in the RBM at positions 479 and 487 determine the progression and the tropism of SARS-CoV-1 [29]. Recent studies using civets, mice and rats demonstrated that any change in these two residues might improve animal-to-human or human-to-human transmission and facilitate efficient cross-species infections. The S2 subunit mediates the fusion between SARS-CoV-1 and target cells. It includes the heptad repeat 1 (HR1) and HR2 domains [29].
3.2. MERS-CoV
Almost ten years after the emergence of SARS-CoV-1, [37] discovered MERS-CoV for the first time in Saudi Arabia in 2012. MERS-CoV has a zoonotic origin and belongs to the Coronaviridae family, Nidovirales order [10]. As of 2016, phylogenetic analysis of MERS-CoV has been done on 182 full-length genomes including 94 humans and 88 from dromedary camels [9]. The MERS-CoV genomes share more than 99% sequence identity, indicating a low mutation rate and low variance among the genomes.
The bat origins of MERS-CoV is supported by evidence that bat coronavirus HKU4 also uses the human MERS-CoV receptor CD26 for virus entry [21,38]. MERS-CoV were found genetically to be highly related to two bat coronaviruses, HKU4 and HKU5 [38]. It is believed that MERS-CoV originated in bats like SARS-CoV-1. According to the WHO, MERS-CoV transmission between humans is possible and occurs in Middle East countries and the Republic of Korea [9]. SARS-CoV-1 was known to cause milder disease, and these outbreaks highlighted their adaptive potential to the changing environmental conditions and they are classified under "emerging viruses" (Figure 3). During MERS-CoV infection, three MERS-CoV proteins are expressed on the envelope of the virus: the surface spike protein (S), the membrane glycoprotein (M) and the envelope protein (E). The virus uses its spike proteins as adhesive factors for host entry through a specific receptor called dipeptidyl peptidase-4 (DPP4 also known as CD26) which is widely expressed on epithelial cells in the kidney, alveoli, small intestine, liver, prostate, and on activated leukocytes [39]. The interaction of MERS-CoVS protein with the DPP4 receptor not only facilitates viral access into the host cell but also triggers signals that induce the immunosuppression of infected patients, enabling viral replication and spread [21,38].
During the infection process, the S protein of MERS-CoV is cleaved into a receptor-binding subunit S1 and a membrane-fusion subunit S2 [40,41]. The MERS-CoV S1 subunit also includes an RBD, mediating the attachment between virus and target cells [41]. The RBD of MERS-CoV is stabilized by three disulfide bonds. The RBM comprises a four-stranded antiparallel β-sheet important for interacting with the extracellular β-propeller domain of DPP4. Regarding a study on cell line susceptibility, MERS-CoV can infect several human cell lines including lower respiratory, kidney, intestinal, and liver cells [42].
3.3. SARS-CoV-2
The novel virus SARS-CoV-2 is a Beta coronavirus, which is enveloped positive-sense RNA, is belong to subgenus sarbecovirus of the Ortho coronavirinae subfamily and is entirely different from the viruses of MERS-CoV and SARS-CoV-1. This virus is encircled with an envelope containing viral nucleocapsid. The electron micrographs of SARS-CoV-2 revealed a diverging spherical outline with some degree of pleomorphism, virion diameters varying from 60 to 140 nm, and distinct spikes of 9 to 12 nm, giving the virus the appearance of a solar corona. RNA genome contains 29891 nucleotides, encoding for 9860 amino acids [43]. Isolated from a COVID-19 patient, the complete genome of SARS-CoV-2, is 29.9 kb [2]. It was found that the genome sequence of SARS-CoV-2 is 79.5%, identical to SARS-CoV-1. While SARS-CoV-1and MERS-CoV have positive-sense RNA genomes of 29.7 kb and 30.1 kb, respectively [44] (Figure 2 and Table 1). The virus genome encodes other structural proteins, including spike (S) protein, envelope (E) protein, matrix (M) protein, and nucleocapsid (N) and several accessory proteins that interfere with the host innate immune response. The M protein is the most abundant viral protein present in the virion, and it binds to the nucleocapsid and acts as a central organizer of coronavirus assembly. The M protein of SARS-CoV-2 does not have an amino acid substitution compared to that of SARS-CoV-1 [31]. The coronavirus E protein plays a multifunctional role in the pathogenesis, assembly and release of the virus [2]. The SARS-CoV-2 E protein reveals a similar amino acid constitution without any substitution [2]. It has noted that N protein plays a role in the formation with the viral genome, facilitates M protein interaction needed during virion assembly, and enhances the transcription efficiency of the virus. Compared to that of SARS-CoV-1, the SARS-CoV-2 N protein possess five amino acid mutations, where two are in the intrinsically dispersed region (IDR; positions 25 and 26), one each in the NTD (position 103), LKR (position 217), and CTD (position 334) [31].
Based on virus genome sequencing results and evolutionary analysis, bat has been suspected as natural host of virus origin via unknown intermediate hosts to infect humans [2] (Figure 3). In the same way, the SARS-CoV-2 shares more genetic similarities with SARS-CoV-1 (79%) and 50% with MERS-CoV [45] (Table 1). Several studies suggested that bat may be the potential reservoir of SARS-CoV-2 [16,45]. After genome sequencing, the SARS-CoV-2was analyzed and showed 96.2% sequence identity with bat-CoV RaTG13, suggesting that bat CoV and human SARS-CoV-2 might share the same ancestor [46]. Both CoVs use the same spike (S) protein for binding to the host cells. Many studies confirmed that the spike protein of SARS-CoV-2 shows a sequence similarity of 76-78% with the spike protein of SARS-CoV-1 [2]. The RBD-S2 shows 73-76% sequence similarity with the S2 domain of the SARS-CoV-1 [47]. Otherwise, the coding regions ofSARS-CoV-2 had a similar genomic organization as that of the bat CoVs and SARS-CoV-1 [47]. The sequence alignment proved that the SARS-CoV-2 genome was related to two viruses originated in bats: bat-SLCoVZC45 (88% identical sequence) and bat-SL-CoVZXC21 (87.5% identical sequence), like the E gene (98.7%), the S gene (75%) of the SARS-CoV-2 [48]. Additionally, most of the SARS-CoV-2 proteins also except for the S proteins (only 80%) and protein 13 (73.2%) Showed sequence similarity with the bat-SL-CoVZC45 and bat-SL-CoVZXC21 [49]. The proteins encoded by SARS-CoV-2, bat-SL-CoVZC45and bat-SL-CoVZXC21 were almost similar in length. The only difference was in S protein of SARS-CoV-2which was longer as compared to the S protein encoded by the bat CoVs, SARS-CoV-1 and MERS-CoV [49,50]. Although, the S2 domain of SARS-CoV-2 showed 93% sequence similarity and S1 showed a sequence similarity of 68% sequence with bat-SL-CoVZC45 andbat-SL-CoVZXC21 S2 and S1 domains, respectively. The receptor-binding domain (S1) of SARS-CoV-2 was closely similar to the S1 domain of SARS-CoV-1 [49-51]. Other structural study has been shown that the genome of SARS-CoV-2 contains a variable number (6-11) of open reading frames (ORFs) [2]. Two-thirds of viral RNA, mainly located in the first ORF (ORF1a/ab) translates two polyproteins, pp1a and pp1ab changes between the early sequence of SARS-CoV-2 and the later isolates of SARS-CoV-1 with nine substitutions in ORF1ab, ORF8 (4 substitutions), the spike gene (3 substitutions), and ORF7a (single substitution) [2]. [48] have recently proved that SARS-CoV-2 contained 16 predicted nspand their functions have been elucidated recently. In the same road, the potential mutation of nsp3 proteins in SARS-CoV-2 could indicate a potential mechanism that differentiates it from other coronavirus. No amino acid substitutions in nsp7, nsp13, envelope, matrix or accessory proteins p6 and 8b, except in nsp2, nsp3, spike protein, underpinning subdomain, i.e., RBD [52]. Another recent research suggested that the mutation in nsp2 and nsp3 play a role in infectious capacity and differentiation mechanism of SARS-CoV-2 [53].
Isolated from the bronchoalveolar lavage fluid (BALF) of a COVID-19 patient, [54] have confirmed that the SARS-CoV-2 uses the same cellular entry receptor ACE2, as SARS-CoV-1 [54]. The ACE2 regulates both the cross-species and human-to-human transmission. The S glycoprotein on the surface of SARS-CoV-2 can attach to the receptor ACE2 on the surface of human cells [2]. The S glycoprotein includes two subunits, S1 and S2. S1 determines the virus-host range and the key function domain-RBD, while S2 mediates virus-cell membrane fusion by two tandem domains, heptad repeats 1 (HR1) and HR2 [50,55]. After membrane fusion, the viral genome RNA is released into the cytoplasm and the RNA translates two polyproteins pp1a and pp1ab, which encode non-structural proteins and form replication-transcription complex (RTC) in double-membrane vesicle. Continuously RTC replicate and synthesize a nested set of sub genomic RNAs, which encode accessory proteins and structural proteins. Mediating endoplasmic reticulum (ER) and Golgi, newly formed genomic RNA, nucleocapsid proteins and envelope glycoproteins assemble and form viral particle buds [56]. Lastly, the virion-containing vesicles fuse with the plasma membrane to release the virus.
Table 1: Summary of biological characteristics of the Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-CoV-1), the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [30].
| EmergingHumanCoronaviruses | |||
|---|---|---|---|
| SARS-CoV-1 | MERS-CoV | SARS-CoV-2 | |
| Starting date | November 2002 | April 2012 | December 2019 |
| First cases | Guangdong, China | SaudiArabia | Wuhan, China |
| Incubation days | 3-7 | 4-6 | 6-14 |
| Reported cases | 8096 | 1905 | >240M (End October 2021) |
| Deaths cases | 774 | 677 | >4.8M (End October 2021) |
| Mortality | More than 10% | More than 35% | More than 3% |
| Affected countries | 29 | 27 | 266 |
| Intermediary host | Civet | Dromaderycamel | Pangolin ??? |
| Transmission regions | globally | regionally | globally |
| Viral replication efficiency | High | High | High |
| Ending date | July 2003 | March 2017 | Not yet |
| symptoms | Fever, cough, fatigue, myalgia, headache | Fever, cough | Fever, cough, fatigue, myalgia, headache, breathing difficulties |
| Specific receptor | Human Angiotensin -Converting Enzyme 2 ACE2 | Human Dipeptidyl Peptidase 4 (DPP4 /CD26) | Human Angiotensin -Converting Enzyme 2 ACE2 |
| RNA Length | 29.727 bases | 30.119 bases | 29.891 bases |
| Open Reading Frames (ORF) | 11 | 11 | 6-11 |
| Structural Protein (sp) | 4 | 4 | 4 |
| Non-Structural Protein (nsp) | 16 | 16 | 16 |
| Spike Protein (nbre of aa) | 1104 | 1206 | 1273 |
| RBD length | 318-513 | 381-588 | 329-521 |
| RBM length | 424-494 | 484-567 | 437-508 |
| HR1 length | 892-1013 | 984-1104 | 912-984 |
| HR2 length | 1145-1195 | 1246-1295 | 1163-1213 |
| Genes order | 5'- replicase ORF1ab, Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N) -3' | ||
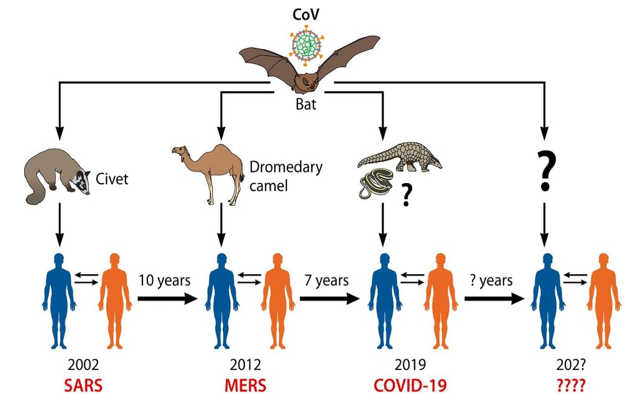
Figure 3: Human CoVs origins. The jumping of wild animal CoVs (bat) to human with intermediate hosts (Spillover infection) is the main origin of emerging human coronaviruses causing outbreaks. The pangolin is unlikely the intermediate animal host of SARS-CoV-2. Attentions should be taken for the next same events in the next periods [1].
4. Emerging human coronaviruses vaccine challenges
Due to the lack of effective antiviral therapy in the present, many attempts are being made to design and develop vaccines for CoV infections. In this part of the review, we discuss therapeutic interventions for CoVs with a special focus on preventive vaccine research strategies. Due to the rapid emergence and spreading of CoVs diseases, efforts toward production an effective vaccine have been developed in many countries. By a lot of knowledge from SARS-CoV-1 and MERS-CoV vaccines development path, several research groups have been able actually to develop many SARS-CoV-2 vaccine candidates.
4.1. SARS-CoV-1
The spike protein S of SARS-CoV-1 is an important and abundant protein, and it is located at outside of virus. Importantly, S protein is dominantly contributed to SARS-CoV-1 specific antibody production, by protein vaccines road and was confirmed to exert the activity by neutralizing SARS-CoV-1. By using SARS-CoV-1 specific antibody (antiserum) and peptide synthesis, four regions were identified in the spike protein with highly immune reactivity67-119aa (I), 265-345aa (II), 588-645aa (III) and 1130-1234aa (IV) respectively [57]. In a study with the DNA vaccine therapy, when the mice were injected with SARS-CoV-1-S-DNA vaccine containing expressible full length of spike gene, anti-ACE-2 binding region antibody was generated and blocking binding of S to ACE-2 [57]. Thus, ACE-2 binding region of spike protein is an antigenic determinant and it may be used as an antigen for SARS-CoV-1vaccination. Other results indicated that DNA vaccine of spike may induce CTL (cytotoxic T lymphocytes) and it is considered as a prospective vaccine candidate for SARS-CoV-1. [58] immunized animals with DNA vaccine containing S gene and obtained high titres of neutralization antibodies capable of blockingSARS-CoV-1 infection [58]. [59] proved that the constructed recombinant forms of S-DNA gene vaccine may elicit protective immunity by a high antibody production anti-SARS-CoV-1-S protein target for vaccination [59]. Recent data demonstrated that ACE-2 is SARS-CoV-1receptor and its ligand is located at region of S1/S2, results indicated that this region contributed to neutralizing antibodies production [57]. Therefore, it is proposed that ACE-2 binding region of SARS-CoV-1spike fragment might contain two antigenic epitopes for neutralizing antibody production and these epitopes might be useful for SARS-CoV-1 vaccination. Consistent with several authors reports that immunization with S1 protein could induce protective immune response [60]. In the same way, [61] used S1 and S2 gene as DNA vaccine and proved that S1 and S2 induced high titres of anti-S1 and anti-S2 antibodies against SARS-CoV-1 spreading [61].
4.2. MERS-CoV
Until now, there are still no commercial vaccines available against MERS-CoV [62]. In MERS-CoV vaccine research, neutralizing antibodies are very potent in neutralizing viral infectivity through blocking MERS-CoV entry into host cells. However, detection of the anti-MERS-CoV antibody response occurs 21 days after infection. It is known that MERS-CoVS spike proteins might be used for blocking the entry of the virus into host cells. A study reported by [63] proved that mice vaccinated with MERS-CoVS protein generate a high level of neutralizing antibodies against homologous viruses [63]. On the other hand, [64] showed that antibodies recovered from the serum of some convalescent SARS-CoV-1 patients might react with MERS-CoV or neutralize it [64]. Full-length spike (S) which contains RBD is considered as a good vaccine antigen because it could prevent host cell attachment and viral entry [63]. Thus, targeting MERS-CoV-1 RBD protein is one of the strategies for vaccine development [65]. The immunogenicity of this fragment within the MERS-CoVS spike protein was tested and evaluated. Remarkably, the 358-588 RBD and 377-662 RBD fragments was shown a higher induction of antibodies [66]. A strong specific IgG antibody response against RBD was generated by MERS-CoVS, 4 to 16 weeks post-immunization and the levels of induced antibodies were significantly higher.
Multiple vaccine candidates targeting the S protein have been developed, including subunit vaccines and DNA vaccines using the DNA vaccine technology [65-66]. Interestingly, DNA vaccine showed the most advance platform in response to emerging pathogens. Similarly, other researchers revealed
that the DNA encoding the S1 domain was demonstrated to be more superior than that encoding the full-length S protein in eliciting antibody and cellular responses. Both DNAs encoding the S1 and S proteins were shown to induce neutralizing antibodies that reacted with MERS-CoV strains of human and camel origins. According to the current technological advancement using mRNA vaccine platform, another nucleic acid-based vaccine has been considered as disruptive vaccine technology and produced humoral and cellular immune responses [66] (summarized in Figure 4). Importantly, the RBD-S-protein induced the highest-titer IgG antibodies in mice [67]. Thus, targeting the interactions between RBD and S protein maybe a potential approach and critical target for neutralizing antibodies. Immunization with N-terminal domain of S1 (NTD) inducing cell-mediated responses in splenocytes as well as humoral responses (IgG) suggest that S1 domains outside the RBD may be used for MERS-CoV vaccines. Actually, none curative or preventive agents is available for clinical use in humans [65].
4.3. SARS-CoV-2
With the global spread of SARS-CoV-2, vaccination must be the most efficient and effective mean to prevent and control COVID-19. Despite the apparent similarity founded between SARS-CoV-1and SARS-CoV-2, there is still considerable genetic variations between viral epitopes. Immune responses against SARS-CoV-1are likely to be effective against SARS-CoV-2 [68]. In fact the potential epitopes become actually a vaccine candidate to developing an effective vaccine against SARS-CoV-2. Recently, a few epitopes have been recognized from the surface of S glycoprotein of SARS-CoV-2. Similar to SARS-CoV-1, SARS-CoV-2 uses spike (S) protein including protein based vaccines technology to gain entry into hostcells. It was shown that the spike (S) protein on the surface of SARS-CoV-2 cell bound the entry receptor angiotensin-converting enzyme 2 (ACE2) on infected cells [2]. Thus, targeting the interactions between ACE2 and S protein may be a potential approach. Specifically, receptor binding domain (RBD) within the S protein is the critical target for neutralizing antibodies. However scientific efforts demonstrate that SARS-CoV-2 spike protein plays a vital role for inducing host immune response and remains a key target for vaccine development and considered as a key target against SARS-CoV-2 infection. By the use of nucleic acid vaccine technology, a potential mRNA vaccine expressing SARS-CoV-2 surface spike protein became a new strategy.
A vaccine against SARS-CoV-2 was urgently needed to prevent further waves and to limit and stop the COVID-19 pandemic. Since the beginning of the SARS-CoV-2 pandemic, more than 160 vaccines candidates are being developed against SARS-CoV-2 by research teams in companies and universities across the world using different biotechnology platforms. On 23rdJanuary, 2020, the Coalition for Epidemic Preparedness Innovations (CEPI) announced the funding to DNA, mRNA, and “molecular clamp” vaccine platforms [69]. The safety of vaccine remains a top priority for vaccine development. From theses vaccines, 137 were undergoing preclinical development and 23 candidates in early clinical development. Researchers were trialing different technologies (Figure 4) some of which haven’t been used in a licensed vaccine before. At least six groups had already begun injecting formulations into volunteers in safety trials; others have started testing in animals. The Figure 4 explains and summarize each vaccine design. All vaccines aim to expose the body to an antigen that won’t cause disease, but will provoke an immune response that can block or kill the virus if a person becomes infected. The body’s adaptive immune system can learn to recognize new, invading pathogens, such as the coronavirus SARS-CoV-2. Within the end of the 2020, there were at least eight types being tried against the coronavirus, and they rely on different viruses or viral parts.
At least seven teams were developing vaccines using the virus itself, in a weakened or inactivated form. Many existing vaccines are made in this way, such as those against measles and polio, but they require extensive safety testing. Sinovac Biotech (Sinopharm company) from the China National Biotech Group in Beijing has developed and tested for the first time an inactivated SARS-CoV-2 vaccine, the BBIBP-CorV in humans at phase 1/2 trial [70]. The inactivated SARS-CoV-2 vaccine, BBIP-CorV, is safe, immunogenic and well tolerated at all tested doses. Rapid humoral responses against the virus were noted from day 4 after the first inoculation and 100% seroconversion was found in all participants on day 42. Two-dose immunization with 4µg
vaccine on days 0 and 21 achieved high neutralizing antibody titers. Mild adverse reactions, including pain and fever, were observed but no severe adverse reaction was reported in all human groups included in the trials. The ongoing phase 1/2 and phase 3 trials provided more information on the safety and immunogenicity, dose and immunization schedule of BBIBP-CorV vaccine. Live-attenuated virus vaccines represent also a potent strategy for viral vaccinations by the injection of the weakened and inactivated SARS-CoV-2. The virus is conventionally weakened for a vaccine by being passed through animal or human cells until it picks up mutations that make it less able to cause disease. Codagenix Company in USA, working with the Serum Institute of India to weaken SARS-CoV-2 by altering its genetic code so that viral proteins are produced less efficiently. They developed a “codon deoptimization” technology to attenuate viruses and is exploring SARS-CoV-2 vaccine strategies [71].
Other developed studies have advanced nucleic acid vaccine for SARS-CoV-2. At least 20 teams were aiming to use genetic instructions (in the form of DNA or RNA) for a coronavirus protein that prompts an immune response. The nucleic acid is inserted into human cells, which then churn out copies of the virus protein; most of these vaccines encode the virus’s spike protein. RNA and DNA based vaccines were safe and easy to develop: to produce them involves making genetic material only, not the virus. Interestingly, Inovio Pharmaceuticals in USA developed a DNA vaccine, such as Moderna Therapeutics, Pfizer and Curevac in USA explored RNA vaccine platforms. Their study evaluated the vaccine’s safety and ability to trigger immune responses. More recently, new formulations have improved nucleic acid performance in humans by the inserted of the genetic material only. This approach eventually lead to the first licensed human nucleic acid vaccine [72].
Many researchers wanted to inject coronavirus proteins directly into the body. Fragments of proteins or protein shells that mimic the coronavirus’s outer coat can also be used. Twenty-eight teams have worked on vaccines with viral protein subunits - most of them are focusing on the virus’s spike protein or a key part of it called the receptor binding domain. Similar vaccines against the SARS-CoV-2 produced by the American vaccine development company Novavax protected monkeys against infection. To work, these vaccines might require adjuvants - immune stimulating molecules delivered alongside the vaccine - as well as multiple doses. Empty virus shells mimic the coronavirus structure but aren’t infectious because they lack genetic material is another new strategy for vaccine development. Five teams were working on “Virus Like Particle” (VLP) vaccines, which can trigger a strong immune response, but difficult to manufacture. AdaptVac that is a joint-venture between NASDAQ and University of Copenhagen spin-out NextGen Vaccines and as part of the EU H2020 funded PREVENT-nCoV consortium, is focused on developing a SARS-CoV-2 vaccine using this VLP display technology.
More than 25 groups worked on viral vector vaccines. A virus such as Measles or Adenovirus is genetically engineered, so that it can produce coronavirus proteins in the body. These viruses are weakened so they cannot cause disease. There are two types of viral vectors, those that can still replicate within cells and those that cannot because key genes have been disabled. The replicating viral vector (such as weakened measles),the newly approved Ebola vaccine is an example of such viral vector vaccine that replicates within cells. Such vaccines were safe and provoked a strong immune response. However, existing immunity to the vector could blunt the vaccine’s effectiveness. The non-replicating viral vector (such as adenovirus),no licensed vaccines use this method in the past, but they have a long history in gene therapy. Using this technology, booster shots are needed to induce long-lasting immunity. According to an industry newsletter, the US-based drug giant Johnson & Johnson was one of the few multinational companies embarking on SARS-CoV-2 vaccines [73] and working on this approach. The ministry of health of the Russian federation designed an heterologous COVID-19 vaccine (Spotnick V) consisting of two components of recombinant adenovirus type 26 (rAd26) vector and a recombinant adenovirus type 5 (rAd5) vector, both carrying the gene SARS-CoV-2 spike glycoprotein (rAd26-S and rAd5-S) suitable for prime-boost vaccination. The vaccine candidate was developed as two formulations (frozen [Gam-COVID-Vac] and lyophilized [Gam-COVID-Vac-Lyo]). According to results obtained from the prime-boost, two open, non-randomized phase 1/2 studies [74], the
heterologous vaccine was safe, well tolerated and does not cause serious adverse events in healthy adults volunteers. The vaccine is highly immunogenic and induce strong humoral and cellular immune responses in 100% of healthy adult volunteers, with antibody titres in vaccinated participants higher than those in convalescent plasma. The phase clinical trial is planned with involvement of 40 000 volunteers from different age and risk groups.
The University of Oxford (Oxford COVID Vaccine Trial Group) with the AstraZeneca company developed and assessed the safety, reactogenicity and immunogenicity of a chimpanzee non replicating adenovirus-vectored vaccine named ChAdOx1 nCoV-19 (AZD1222) vaccine expressing the SARS-CoV-2 spike protein [75]. Results of the first clinical phase 1/2, single-blind and randomized controlled trial of this vaccine demonstrated that ChAdOx1 nCoV-19 was safe, tolerated and immunogenic with reduced reactogenicity when paracetamol was used prophylactically for the 24 h after vaccination. A single dose elicited both humoral and cellular responses against SARS-CoV-2, with a booster immunisation augmenting neutralizing antibody titres. The ChAdOx1 nCoV-19 vaccine should be used to prevent infection, disease and death in the global population, with high-risk populations such as hospital workers and older adults priorotised to receive vaccination. However, the immune correlates of protection against SARS-CoV-2 have not yet been determined. The same strategy and the same platform technology was used by researchers from Beijing Institute of Biotechnology in China in collaboration with CanSino Biologics company [76]. The results of randomized controlled trial to assess the immunogenicity and safety of a candidate non replicating adenovirus type 5 (Ad5)-vectored COVID-19 vaccine demonstrated that the vaccine has a good safety profile, a single-dose immunisation induce rapid onset of immune responses within 14 days and significant humoral and cellular responses within 28 days. Such results support testing of the Ad5-vectored COVID-19 vaccine at dose of 5 x 1010 viral particles in programmed international multicenter, randomized, double-blind, controlled phase 3 effectiveness trial in healthy adults to further evaluate the efficacity of the vaccine.
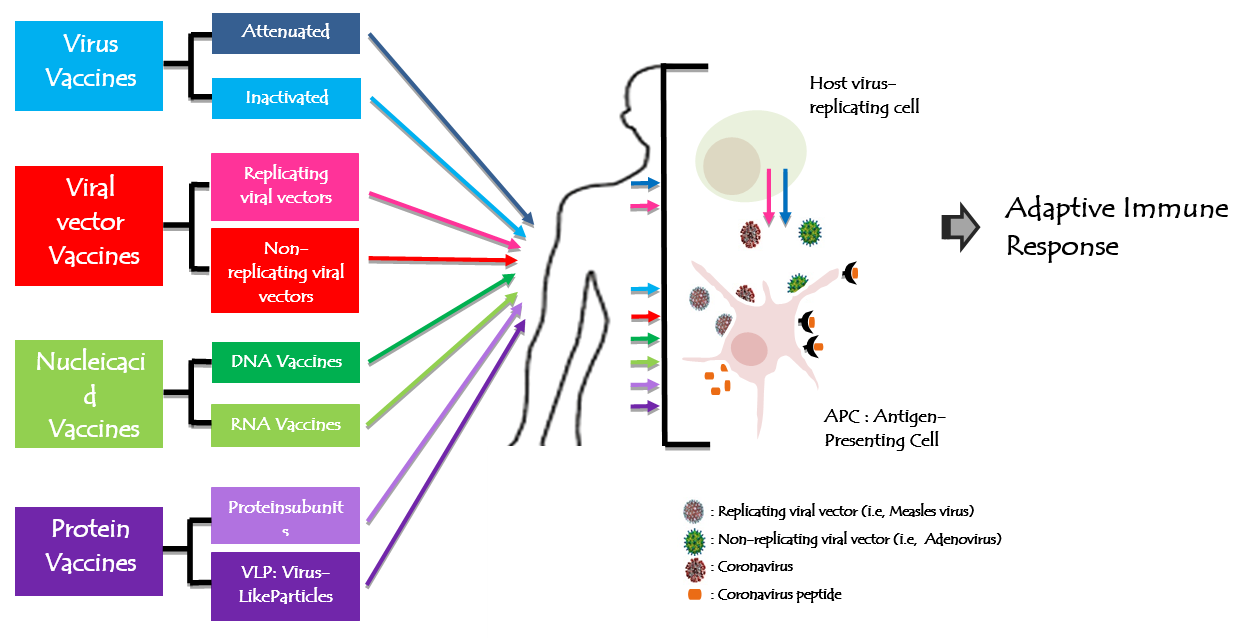
Figure 4: Figure 4: Human coronavirus vaccine technologies. All vaccines aim to expose the body to an antigen that won't cause disease and will provoke an adaptive immune response against the virus. Actually, there are 8 types of biotechnology platforms developing vaccines tried in different trial steps.
5. Conclusion
SARS-CoV-2 pandemic has a significant global impact and challenged the social, economic, medical, cultural and public health infrastructure of the entire world that may take many years to recover. Many antiviral therapies strategies based on the updated research data for CoVs and their emergences are currently evaluated. Among these strategies ways, we suppose that vaccines are the best way for the limitation of
SARS-CoV-2 spread and the other emerging human coronaviruses. Therefore, scientific efforts should be made to devise comprehensive measures to help in design prophylactic and therapeutic measures for future outbreaks of similar coronaviruses jumping from wild animal especially bats to human.
Conflict of interest: All authors have no conflict of interest.
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
6. References
- Kuldeep D, Sharun K, Ruchi,T, Shubhankar S, Sudipta B, et al. (2020) Coronavirus Disease 2019-COVID-19. Clin Microbiol Rev. 33(4): e00028-20. [PubMed.]
- Manisha Y, Swasti D,Satya E, (2020)Emerging strategies on in silico drug development against COVID-19: challenges and opportunities. Eur J Pharm Sci. 155: 1-16. [Ref.]
- Chang CK, Lo SC, Wang YS, Hou MH. (2016) Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov Today. 21: 562-72. [PubMed.]
- Wang W, Xu Y, Gao R, Lu R, Han K, et al. (2020) Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 323(18): 1843-1844. [Ref.]
- Tanu S. (2020) A Review of Coronavirus Disease-2019 (COVID-19). Indian J Pediatr. 87(4): 281-286. [PubMed.]
- Saif LJ. (2004) Animal coronaviruses: What can they teach us about the severe acute respiratory syndrome? Rev Sci Tech. 23: 643-60. [PubMed.]
- Lee N, Hui D, Wu A, Chan P, Cameron P,et al (2003). A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 348: 1986-1994. [PubMed.]
- Al-Osail AM, Al-Wazzah MJ. (2017). The history and epidemiology of Middle East respiratory syndrome corona virus. MultidiscipRespirMed. 12: 20. [PubMed.]
- Enjuanes L, Zuñiga S, Castaño-Rodriguez C, Gutierrez-Alvarez J, Canton J, et al. (2016) Molecular Basis of Coronavirus Virulence and Vaccine Development. Adv Virus Res. 96: 245-286. [PubMed.]
- Ayman M, Wael A, Maged GH. (2019) Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Infection Immunological Response and Vaccine Development.J Immunol Res. 7: 6491738. [Ref.]
- Hijawi B, Abdallat M, Sayadeh A, Alqasrawi S, Haddadin A. (2012) From a retrospective investigation Novel coronavirus infection in Jordan. epidemiological finding EMHJ. 19(1): S12-18. [PubMed.]
- Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, et al. (2012) Severe respiratory illness caused by a novel coronavirus in a patient transferred to the United Kingdom from the Middle East September 2012. Euro Surveill. 17: 40. [Ref.]
- Buchholz U, Müller MA, Nitsche SA, Wevering N, Bauer-Balci, et al. (2012) Contact investigation of a case of human novel coronavirus infection treated in a German hospital October–November 2012. Euro Surveill. 21: 18(8), 20406. [PubMed.]
- Gulland A. (2013) Novel coronavirus spreads to Tunisia. BMJ. 346. [PubMed.]
- Puzelli S, Azzi A, Santini MG, Martino D, A Facchini, et al. (2013) Investigation of an imported case of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) infection in Florence Italy May to June 2013. Euro Surveill. 18(34): 20564. [Ref.]
- Chan J, Yuan S, Kok K, To K, Chu H, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 395 (10223): 514-523. [PubMed.]
- Yuen K, Ye Z, Fung S, Chan C, Yin D (2020) SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 10: 40. [Ref.]
- Chan JF, Yuan S, Kok KH, To KK, Chu H, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 395:514-523. [Ref.]
- Yang L, Peng H, Zhaoling Z, Gang L, Zitong H, et al. (2007) Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J GenVirol. 88(10):2740-2748. [PubMed.]
- McBride R, Van Zyl M, Fielding BC. (2014) The coronavirus nucleocapsid is a multifunctional protein. Viruses. 6:2991-3018. [Ref.]
- Li F. (2016) Structure function and evolution of coronavirus spike proteins. Annu Rev Virol. 3:237-61. [PubMed.]
- Guo Y, Korteweg C, McNutt MA, Gu J. (2008) Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 133: 4-12. [PubMed.]
- Te Velthuis AJ, Van den Worm SH, Snijder EJ. (2012) The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 40: 1737-47. [Ref.]
- Stobart CC, Sexton NR, Munjal H, Lu X, Molland KL, et al. (2013) Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. J Virol. 87: 12611-12648 [Ref.]
- Wang H, Xue S, Yang H, Chen C. (2016) Recent progress in the discovery of inhibitors targeting coronavirus proteases. Virol Sin. 31: 24-30. [PubMed.]
- Krichel B, Bylapudi G, Schmidt C, Blandet C, Shubert R, et al. (2021) Hallmarks of Alpha- and Betacoronavirus non-structural protein 7+8 complexes. Sci Adv. 3: 7(10). [Ref.]
- Snijder EJ, Decroly E, Ziebuhr J. 2016 The nonstructural proteins directing Coronavirus RNA synthesis and processing. Adv Virus Res. 96: 59-126. [PubMed.]
- Bouvet M, Lugari A, Posthuma CC, Zevenhoven JC, Bernard S, et al. (2014) Coronavirus Nsp10 a critical co-factor for activation of multiple replicative enzymes. J Biol Chem. 289: 25783-25796. [Ref.]
- Li F, Li W, Farzan M, Harrison SC. (2005) Structure of SARS coronavirus spike receptor-binding domain complexed with receptor Science 309: 1864-1868. [PubMed.]
- Zhiqi S, Yanfeng X, Linlin B, Ling Z, Pin Y, et al. (2019) From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 11(59):1-28. [PubMed.]
- Chang MS, Lu YT, Ho ST, Wu CC, Wei TY, et al. (2004) Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem Biophys Res Commun. 314:931-936. [PubMed.]
- Rachel U, Isabelle I, Bruno C, John Z . (2009) Expression and Functions of SARS Coronavirus Replicative Proteins Molecular Biology of the SARS-Coronavirus. PMC 22: 75-98. [Ref.]
- Ito N E C, Mossel K, Narayanan VL, Popov C, Huang T, et al. (2005) Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J Virol. 79:3182-3186. [PubMed.]
- Jonathan K. (2003) Is the SARS virus mutating? Nature. 10:1. [Ref.]
- Ding Y, He L, Zhang Q, Huang Z, Che X, et al. (2004) Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: Implications for pathogenesis and virus transmission pathways. J Pathol. 203: 622-630. [PubMed.]
- Marco AM, Steven JM, Jones CR, Astell RA, Holt AB, et al. (2003) The Genome Sequence of the SARS-associated Coronavirus. Science. 300(5624): 1399-1404. [PubMed.]
- Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 367: 1814-1820. [PubMed.]
- Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, et al. (2014) Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. BiochemBiophys Res Commun. 451: 208-214. [PubMed.]
- Thea M, Manrico M, Emilio J. 2020 Focus on Receptors for Coronaviruses with Special Reference to Angiotensin-converting Enzyme 2 as a Potential Drug Target - A Perspective. EndocrMetab Immune Disord Drug Targets. 20(6): 807-811. [PubMed.]
- Stalin R, Huihui M, Saskia LS, Dick H, Dekkers W, et al. (2013) Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC Nature 495(7440), 251-254. [PubMed.]
- Zhang N, Jiang S, Du L. (2014) Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines. 13(6): 761-774. [PubMed.]
- Du L, Zhao G, Kou Z, Cuiqing M, Shihui S, et al. (2013) Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol. 87(17): 9939-9942. [PubMed.]
- Jasper FW, Chan KH, Kok Z, Zhu H, Chu KKW, et al. (2020) Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 9(1): 221-236. [Ref.]
- Yang H, Bartlam M, Rao Z. (2006) Drug design targeting the main protease the Achilles' heel of coronaviruses. Curr Pharm Des. 12: 4573-90. [PubMed.]
- Lu R, Zhao X, Li J, Peihua N, Bo Y, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 395: 565-574. [Ref.]
- Ji W, Wang W, Zhao X, Zai J, Li X. (2020) Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J Medi Virol. 92:4. [PubMed.]
- Ali A, Rabaan SH, Al-Ahmed H, ShafiulH, RanjitS, et al. (2020) SARS-CoV-2 SARS-CoV and MERS-CoV: a comparative overview. Le Infezioni in Medicina. 174-184. [PubMed.]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature. 579(7798): 265-269. [PubMed.]
- Jiang S, Du L, Shi Z. (2020) An emerging coronavirus causing pneumonia outbreak in Wuhan China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect. 9: 275-277. [PubMed.]
- Wan Y, Shang J, Graham R, Baric RS, Li F. (2020) Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. JVirol. 94(7): 127-20. [PubMed.]
- Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, et al. (2014) Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio5. e00884-14. [Ref.]
- Aiping W, Yousong P, Baoying H, Xiao D, Xianyue W, et al. (2020) Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host & Microbe. 27 (3): 325-328. [Ref.]
- Domenico B, Martina BM, Giovanetti S, Pascarella MC. (2020) COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis. J Medi Virol. 92 :584-588. [PubMed.]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579: 270-273. [PubMed.]
- Fei Y, Lanying D, David MO, Chungen P, Shibo J. (2020) Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan. ChinaMicrobes Infect. 22(2): 74-79. [Ref.]
- Perrier AB, Desmarets L, Danneels A, Goffard A, Rouille Y. (2019) The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 294(39): 14406-14421. [PubMed.]
- Minsheng Z. (2004) SARS Immunity and Vaccination.Cellul Molecul Immunol. (1): 3. [PubMed.]
- Yang ZY, Kong WP, Huang Y. (2004) A DNA va-ccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 428:561-564. [Ref.]
- Bisht H, Roberts A, Vogel L. (2004) severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl AcadSci USA. 101: 6641-6646. [PubMed.]
- Johnson MA, Pooley C, Ignjatovic J, Tyack SG. (2003) A recombinant fowl adenovirus expressing the S1 gene of infectious bronchitis virus protects against challenge with infectious bronchitis virus. Vaccine. 21: 2730-2736. [PubMed.]
- Zeng F, Chow KY, Hon CC. (2004) Characterization of humoral responses in mice immunized with plasmid DNAs encoding SARS-CoV spike gene fragments. BiochemBiophysRes Commun. 315: 1134-1139. [PubMed.]
- Welsh RM, Waggoner SN. (2013) NK cells controlling virus-specific T cells: rheostats for acute vs persistent infections. Virology. 435(1): 37-45. [PubMed.]
- Coleman CM, Liu YV, Mu H, Justin KT, Michael M, et al. (2014) Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 32(26): 3169-3174. [PubMed.]
- Chen J, Subbarao K. (2007) The immunobiology of SARS. Annu Rev Immunol 25: 443-472. [PubMed.]
- Tang J, Zhang N, Tao X, Guangyu Z ,Yan G, et al. (2015) Optimization of antigen dose for a receptor-binding domain-based subunit vaccine against MERS coronavirus. Human Vaccines Immunotherapeutics. 11(5): 1244-1250. [PubMed.]
- Mou H, Raj VS, Van Kuppeveld FJM, Rottier PJM, Haagmans BL, et al. (2013) The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol. 87(16): 9379-9383. [PubMed.]
- Du L, Zhao G, He Y, YanGuo BZ, Shibo J, et al. (2007) Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 25(15): 2832-2838. [PubMed.]
- Syed FA, Ahmed AQ, Matthew RM. (2020) Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 12(3): 254. [RePubMedf.]
- Pang J, Wang MX, Ang IYH, Tan SHX, Lewis RF, et al. (2020) Potential rapid diagnostics vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J Clin Med. 9(3): 623. [Ref.]
- Shengli X, Yuntao Z, Yanxia W, Hui W, Yunkai Y, et al. (2020) The Lancet Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. 21(1): 39-51. [PubMed.]
- Shieber J (2020) Codagenix raises $20 million for a new flu vaccine and other therapies Tech. [Ref.]
- Inovio Pharmaceuticals (2020) Inovio Collaborating with Beijing Advaccine to Advance INO-4800 Vaccine Against New Coronavirus In China. [Ref.]
- Wen-Hsiang C, Ulrich S, Peter JH, Maria EB. (2020) The SARS-CoV-2 Vaccine Pipeline: An Overview Curr. Trop Med Rep 3:1-4. [PubMed.]
- Denis YL, Inna VD, Olga VZ, Amir IT, Dmitry VS, et al. (2020) Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. The Lancet. 395(10240):1845-1854. [PubMed.]
- Pedro MF, Katie JE, Parvinder KA, Brian A, Steph an B, et al. (2020) Safety and immunogenicity of th e ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, singleblind, randomised controlled trial Lancet.396(10249): 467-478. [PubMed.]
- Feng-Cai Z, Xu-Hua G, Yu-Hua L, Jian-Ying H, Tao J, et al. (2020) Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. The lancet. 396(10249): 479-488. [Ref.]